Background: Several recent studies support the notion of excessive complement activation in patients with severe coronavirus disease-19 (COVID-19), with beneficial results of complement inhibition in case series. In this context, severe COVID-19 shares common characteristics with complement-mediated thrombotic microangiopathy (TMA). TMA is commonly characterized by genetic susceptibility, and presents with thrombocytopenia, anemia, increased lactate dehydrogenase (LDH), and organ damage (renal, neurological, cardiac).
Aims: We hypothesized that genetic susceptibility would be also evident in patients with severe COVID-19 and would be associated with disease severity.
Methods: We prospectively studied consecutive adult patients hospitalized with COVID-19 in our referral centers (April-May 2020). Diagnosis was confirmed by reverse-transcriptase polymerase chain reaction (RT-PCR). COVID-19 severity was assessed based on World Health Organization's (WHO) criteria into moderate/severe, and critical disease. Additional data on patients' history and course were recorded by treating physicians that followed patients up to discharge or death. Patients' DNA was obtained from peripheral blood samples. Probes were designed using the Design studio (Illumina). Amplicons cover exonic regions of TMA-associated genes (Complement factor H/CFH, CFH-related, CFI, CFB, CFD, C3, CD55, C5, MCP, thombomodulin/THBD, ADAMTS13) spanning 15 bases into the intronic regions. We used 10ng of initial DNA material. Libraries were quantified using Qubit and sequenced on a MiniSeq System in a 2x150 bp run. Analysis was performed using the TruSeq Amplicon application (BaseSpace). Alignment was based on the banded Smith-Waterman algorithm in the targeted regions (specified in a manifest file). We performed variant calling with the Illumina-developed Somatic Variant Caller in germline mode and variant allele frequency higher than 20%. Both Ensembl and Refseq were used for annotation of the output files. Variants clinical significance was based on ClinVar and the current version of the Complement Database, as we have previously described.
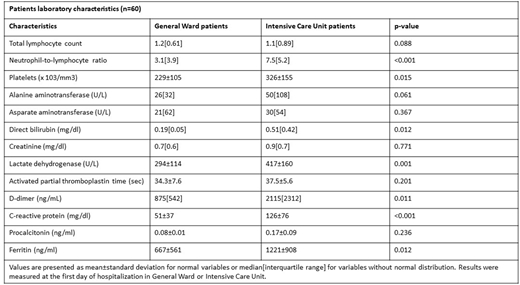
Results: We studied 60 patients, 40 with moderate/severe disease hospitalized in COVID-19 general ward (GW) and 20 with critical disease hospitalized in intensive care units (ICU). Among them, 11 patients succumbed due to COVID-19 disease. Patients laboratory characteristics are shown in Figure.
In genetic analysis, patients presented heterogeneous variant profiles including pathogenic, benign, likely benign, and variants of unknown significance (median number of variants: 62, range: 51-89). Search in the Complement Database revealed seven patients, each carrying one pathogenic or likely pathogenic variant in C3, CD46, DGKE, and CFH. Based on ClinVar, we found a pathogenic variant of ADAMTS13 (rs2301612, missense) in 28 patients. We also detected two missense risk factor variants, previously detected in complement-related diseases: rs2230199 in C3 (13 patients); and rs800292 in CFH (26 patients). Among them, 22 patients had a combination of these characterized variants. This combination was significantly associated with critical disease that required intensive care (p=0.037), as well as low lymphocyte counts (p=0.021) and high neutrophil-to-lymphocyte ratio (p=0.050). In the multivariate model, critical disease was an independent predictor of double heterozygocity in these variants. Furthermore, one patient had a rare germline missense variant in CFI (rs112534524), previously detected in complement-related diseases. This patient suffered from critical disease but survived after long-term ICU hospitalization. Interestingly, five patients showed a likely protective missense variant in CFB (rs641153).
Conclusion: We have detected for the first time rare and pathogenic TMA-associated variants in patients with severe COVID-19. Our findings of variants in complement-regulatory genes and ADAMTS13 suggest genetic susceptibility and define proof-of-concept for proper selection of patients that would benefit from complement inhibition.
Gavriilaki:Omeros Pharmaceuticals: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal